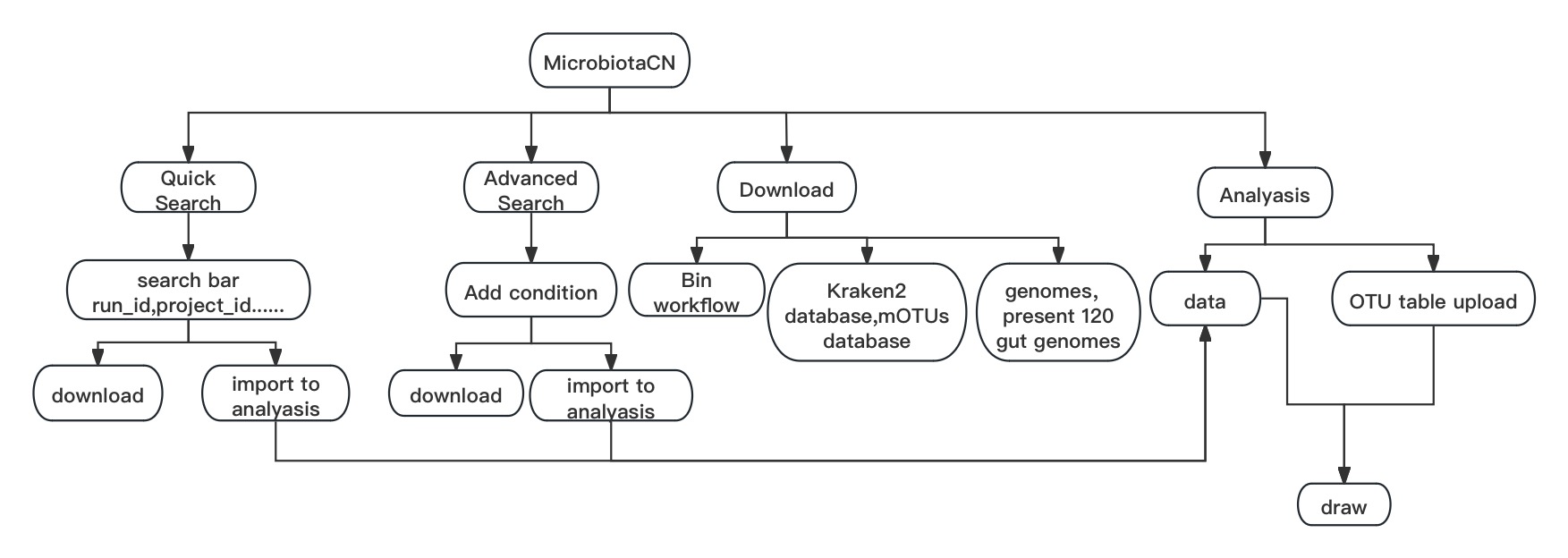
MicrobiotaCN
Search human gut metagenomic metadata and profile, and download them of selected entries. Analyze with them or your own dataset.
Quick Search
The user can filter the entries by using the available main filters or search box in the “Quick search” section. After filtering, the user can download metadata and profile of selected entries, or import them to “Analyze” section.
Advanced Search
The “Advanced search” section provides the user with the ability to search with more complex conditions but slowlier. The user can download metadata and profile of selected entries, or import them to “Analyze” section, too.
Download
The user can download the database and tools in the “Download” section to obtain profiles of their own metagenomic data. And then the users can analyze them to “Analyze” section in isolation or with other data download from Search section.
Analyze
“Analyze” section provides an interactive way of bioinfomatic analysis online with metadata and profile filtered in search page or uploaded by the user. The user can plot with interactive options and download images.
Tutorial video
Help
1.Home
The MicrobiotaCN user interface is divided into three main sections so users can choose the section that better fits their needs. The first section, “Quick search”, holds the full content of the databases' current version, as well as the ability to filter the samples through their main characteristics. The “Advanced search” section provides the user with the ability to search with more complex conditions but slowlier. The “Download” section provides the user with our genomes collection and the Kraken2 and mOTUs database built by our genomes collection. Finally, the “Analyze” section provides an interactive way of bioinfomatic analysis online with metadata and profile filtered in search page or uploaded by the user.
2. How to use “Quick Search”?
“Quick search” is available for users to access the full content of HumanMetagenomeDB, as well as filter the dataset according to the main available features.
Example usage:
- All metagenomes metadata will be shown in the Quick search section, including the ones without valid coordinates.
- You can filter the entries by using the available main filters or by typing in the search box that is placed at the top of the table.
- After filtering, you can download the metadata of your selected entries as a tab-separated values (.txt) file.
- At the same time, you can preview and download the profile of your selected entries as a tab-separated values (.txt) file, too.
- You can import the metadata and profile of your selected entries into “Analyze” by clicking “Import to Analyze” buttom after login in “Analyze” section.
3. How to use “Advanced search?”
“Advanced search” section provides to the user the ability to search with more complex conditions but slowlier, especillay when uses the condition of “Equals”. It is recommand to use other condition like “Contains” to speedup
Example usage:
- All metagenomes metadata will be shown in the Quick search section, including the ones without valid coordinates.
- You can filter the entries by using the Custom Search Builder at the top of the table by “Add contidion”.
- After filtering, you can download the metadata of your selected entries as a tab-separated values (.txt) file.
- At the same time, you can preview and download the profile of your selected entries as a tab-separated values (.txt) file, too.
- You can import the metadata and profile of your selected entries into “Analyze” by clicking “Import to Analyze” button after login in “Analyze” section.
4. How to download genomes collection?
“Download” section provides the user with our genomes collection. Users can download our genomes collection by downloading MBCN_genomes, Or download streamlined genomes of 120 bacteria(MBCN_bac120).
5. Taxonomic assignment to metagenomic reads?
“Download” section provides the user with the Kraken2 and mOTUs database built by our genomes collection. Users can download the database and software to obtain the profiles of their own metagenomic data. It's recommended that you use the Kraken2 database when you have enough computing resource. Only when your server is lack of memory, the mOTUs database is recommended
6. How to use “Analyze”?
“Analyze” section provides an interactive way of bioinfomatic analysis online with metadata and profile filtered in search page or uploaded by the user.
Example usage:
- First, login “Analyze” section.
- Click “Import to Analyze” button in search page after filtering to upload profile and metadata, and then begin to plot.
- You can upload your own profile in “OTU table” tab and metadata in “Factor metadata” and “Numeric metadata” tab instead.
- When you upload your own profile, the first column should be taxonomy name with 7 levels split by '.', ',', ';' or '|'. For example, 'k__Bacteria;p__Actinobacteria;c__Actinobacteria;o__Bifidobacteriales;f__Bifidobacteriaceae;g__Bifidobacterium;s__Bifidobacterium_longum'. Otherwise, you must uploaded an OTUtotax file which contains the 7 levels of the taxonomy in 'OTU to tax table' Page like example. The first column of OTUtotax table should match the first column of profile.
- When you upload your own metadata, the first column of metadata should be sample ids, which must match column names of OTU table. The factor metadata is necessary for all analysis. The numeric metadata is only needed in “Environmental” analysis tab.
- You can skip all upload steps and begin analysis with example data.
- Click “Draw” button to plot and “Download” button to download the plot. If “Download” button doesn't work, you can try to right-click the image of plot to save as png file.
7. What does each attribute of the database mean?
8. Other questions?
Please contact with us by email aetherwalker965@gmail.com